Bio-Inspired Hashing for Unsupervised Similarity Search
Authors
Authors
- Chaitanya K. Ryali
- John J. Hopfield
- Leopold Grinberg
- Dmitry Krotov
Authors
- Chaitanya K. Ryali
- John J. Hopfield
- Leopold Grinberg
- Dmitry Krotov
Published on
01/14/2020
Categories
What can a fruit fly teach us about machine learning? Quite a bit, it turns out. The common fruit fly can recognize and categorize many different odors, and this drives much of the insect’s behavior. In our paper, Bio-Inspired Hashing for Unsupervised Similarity Search, published in ICML 2020, we use the inspiration from the fruit fly olfactory network and a biologically plausible method for representation learning to propose a data-driven hashing algorithm for approximate similarity search.
Combining sparse expansive networks with bio-plausible learning
Sparse expansive representations are ubiquitous in neurobiology. Expansion means that a high-dimensional input is mapped to an even higher dimensional secondary representation. Such expansion is often accompanied by a sparsification of the activations: dense input data is mapped into a sparse code, where only a small number of secondary neurons respond to a given stimulus. This network motif is shown in Figure 1 (below), where a dense high dimensional input vector, is mapped into a sparse binary vector of even higher dimensionality. A classic example of the sparse expansive motif is the fruit fly olfactory system, where approximately 50 projection neurons send their activities to about 2500 Kenyon cells (Turner, et al.). A similar motif can be found in the olfactory circuits of rodents (Mombaerts, et al.), and in the cerebellum and hippocampus of rats (Dasgupta, et al.).
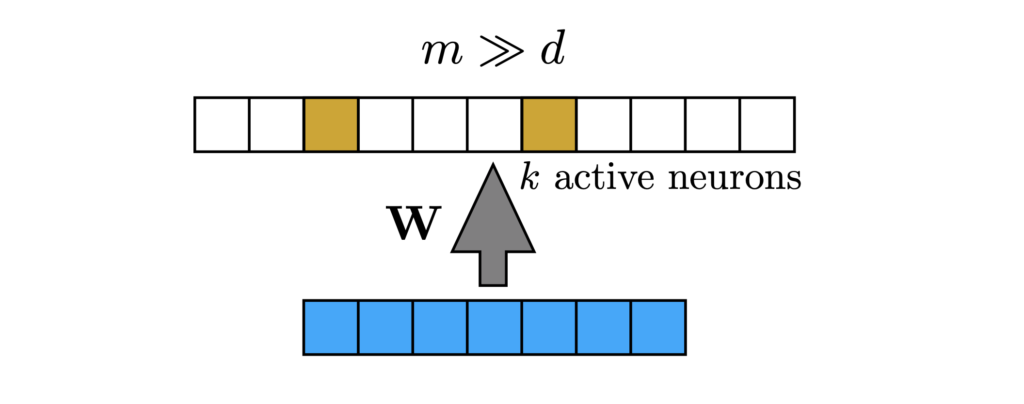
Figure 1: Sparse expansive network motif. Large dimensional input (of dimension ![]() ) is mapped into an even larger dimensional latent space (of dimension
) is mapped into an even larger dimensional latent space (of dimension ![]() ). The projections can be random or data driven.
). The projections can be random or data driven.
Similarity search is a fundamental problem in computer science. Given a query item (for example an image) and a database containing many similar items, the task is to retrieve a ranked list of items from the database most similar to the query. When data is high-dimensional (e.g. images/documents) and the databases are large, this is a computationally challenging problem. However, approximate solutions are generally acceptable, with Locality Sensitive Hashing (LSH) being one such approach. In LSH, the idea is to encode each database entry with a sparse binary vector and to retrieve a list of entries that have the smallest Hamming distances with the query.
Inspired by the fruit fly’s sparse expansive motifs, a new family of hashing algorithms has been recently proposed (Dasgupta, et al.). These algorithms, however, use random weights to accomplish the expansion in the representational space and cannot learn from the data.
In our work, we combine this sparse expansive network motif with the bio-inspired learning algorithm (Krotov & Hopfield), motivated by local Hebbian-like plasticity, to design a novel hashing algorithm called BioHash. The key difference of our algorithm relative to the one proposed in (Dasgupta, et al.) is that the synaptic weights in our networks are learned from the data, rather than being random. We demonstrate that BioHash significantly improves retrieval performance on common machine learning benchmarks.
In Tables 1 and 2 one can see the mean averaged precision (mAP) for retrievals on the MNIST and CIFAR-10 datasets.
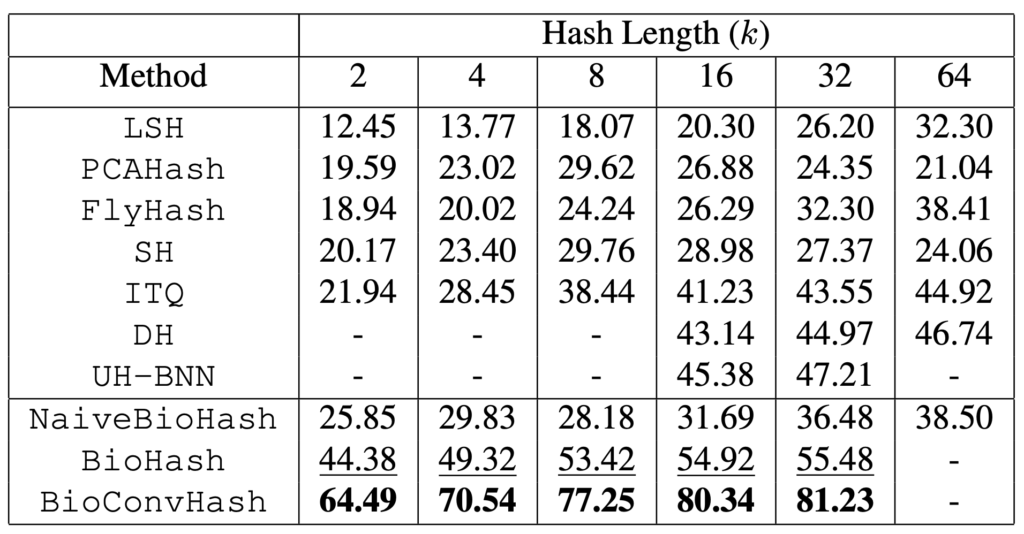
Table 1: mAP@All (%) on MNIST (higher is better). Best results (second best) for each hash length are in bold (underlined). BioHash demonstrates the best retrieval performance, substantially outperforming other methods including deep hashing methods DH and UH-BNN, especially at small hash lengths. Performance for DH and UH-BNN is unavailable for some hash lengths, since it is not reported in the literature.
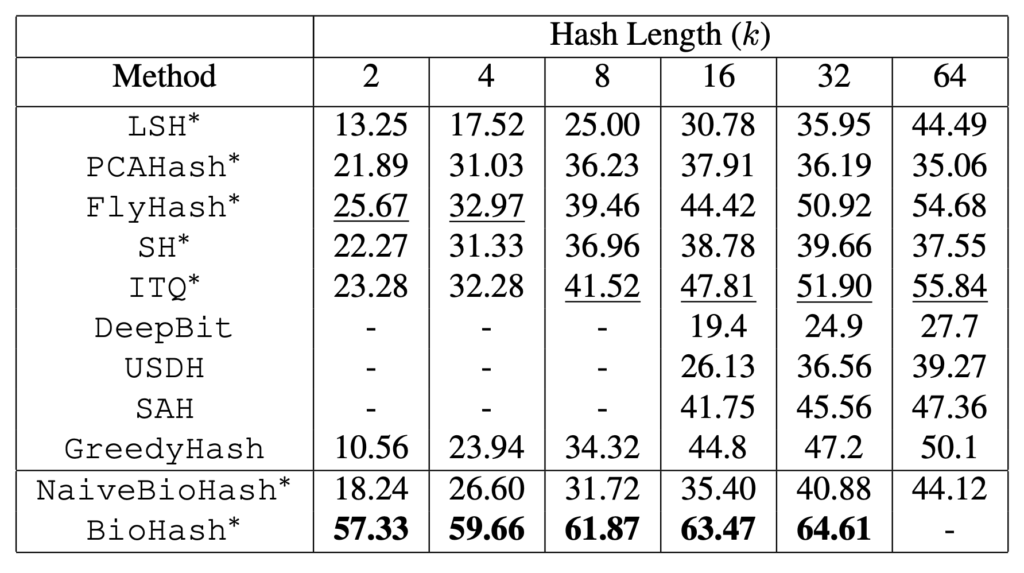
Table 2: mAP@1000 (%) on CIFAR-10CNN. Best results (second best) for each hash length are in bold (underlined). BioHash demonstrates the best retrieval performance, substantially outperforming other methods including deep hashing methods GreedyHash, SAH, DeepBit and USDH, especially at small hash lengths. Performance for DeepBit, SAH and USDH is unavailable for some hash lengths, since it is not reported in the literature.
It is clear that our algorithms (BioHash and its convolutional version BioConvHash) demonstrate a significantly higher values of mAP compared to previously published benchmarks. For instance, we demonstrate that for certain hash lengths BioHash results in approximately 3x improvement in the mean average precision of the retrievals compared to previously published algorithms. In Figure 2 we show examples of queries and corresponding retrievals generated by our method (first four rows are examples of good retrievals, last two rows are examples of bad retrievals).
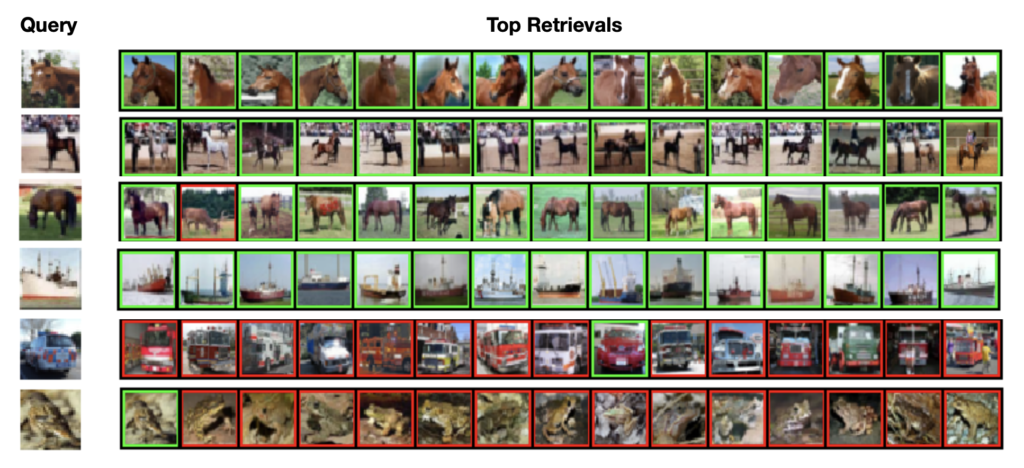
Figure 2. Examples of queries and top 15 retrievals using BioHash (k = 16) on VGG16 fc7 features of CIFAR-10. Retrievals have a green (red) border if the image is in the same (different) semantic class as the query image. We show some success (top 4) and failure (bottom 2) cases. However, it can be seen that even the failure cases are reasonable.
The intuition behind the BioHash learning algorithm is this: we can think about a hidden unit’s synaptic weights as a particle that moves in the space of the data under the influence of two forces: the force of attraction to the peaks of the data distribution, and the force of repulsion between the hidden units.
The learning process is described as a collective motion of many particles (hidden units) each experiencing these two forces. The steady state distribution of the hidden units is determined by the balance of these two forces. Intuitively, this is a useful computational strategy since we only need hidden units receiving the inputs from the regions of the input space where there is a non-zero density of the data. This is accomplished by the force of attraction to the data. At the same time, given a certain number of hidden units they need to be distributed in the space in such a way so that the entire support of the data distribution is covered. This is accomplished by the force of repulsion between the hidden units.
What we discussed above is an example of how biological inspiration can help us achieve significant performance gains in machine learning. An opposite perspective on these results (“an idea for neuroscience from AI”), already discussed (Dasgupta, et al.), is the proposal that Locality Sensitive Hashing might be the computational role of sparse expansive network motifs in biology. Our work provides the existence proof of this proposal in the context of synaptic weights learned in a neurobiologically plausible way.
Note: this post describes a research algorithm nicknamed BioHash, not to be confused with Biometric BioHASH® SDK by GenKey. The name BioHash in this post is used for clarity relative to the research publication and does not refer to any IBM products.
Please cite our work using the BibTeX below.
@misc{ryali2020bioinspired,
title={Bio-Inspired Hashing for Unsupervised Similarity Search},
author={Chaitanya K. Ryali and John J. Hopfield and Leopold Grinberg and Dmitry Krotov},
year={2020},
eprint={2001.04907},
archivePrefix={arXiv},
primaryClass={cs.LG}
}